Infectious Diseases (INF)
Poster Session II
PII-024 - TRANSLATIONAL MODEL-BASED META-ANALYSIS - A POWERFUL QUANTITATIVE TOOL IN DEVELOPING VACCINES AGAINST SARS-COV-2 VARIANTS.
Thursday, March 23, 2023
5:00 PM - 6:30 PM EDT
N. Plock1, B. Kandala2, A. Largajolli1, K. Watson1, R. Thatavarti1, S. Cheung1, J. Sachs2; 1Certara, Princeton, NJ, USA, 2Merck & Co., Rahway, NJ, USA.
.jpg)
Bhargava Kandala, PhD
Director
Merck & Co.
Bridgewater, New Jersey, United States
Presenting Author(s)
Background: Quantitative models leveraging non-clinical data to predict clinical vaccine efficacy provide a translational framework to rapidly develop vaccines/boosters against new strains of SARS-CoV-2.
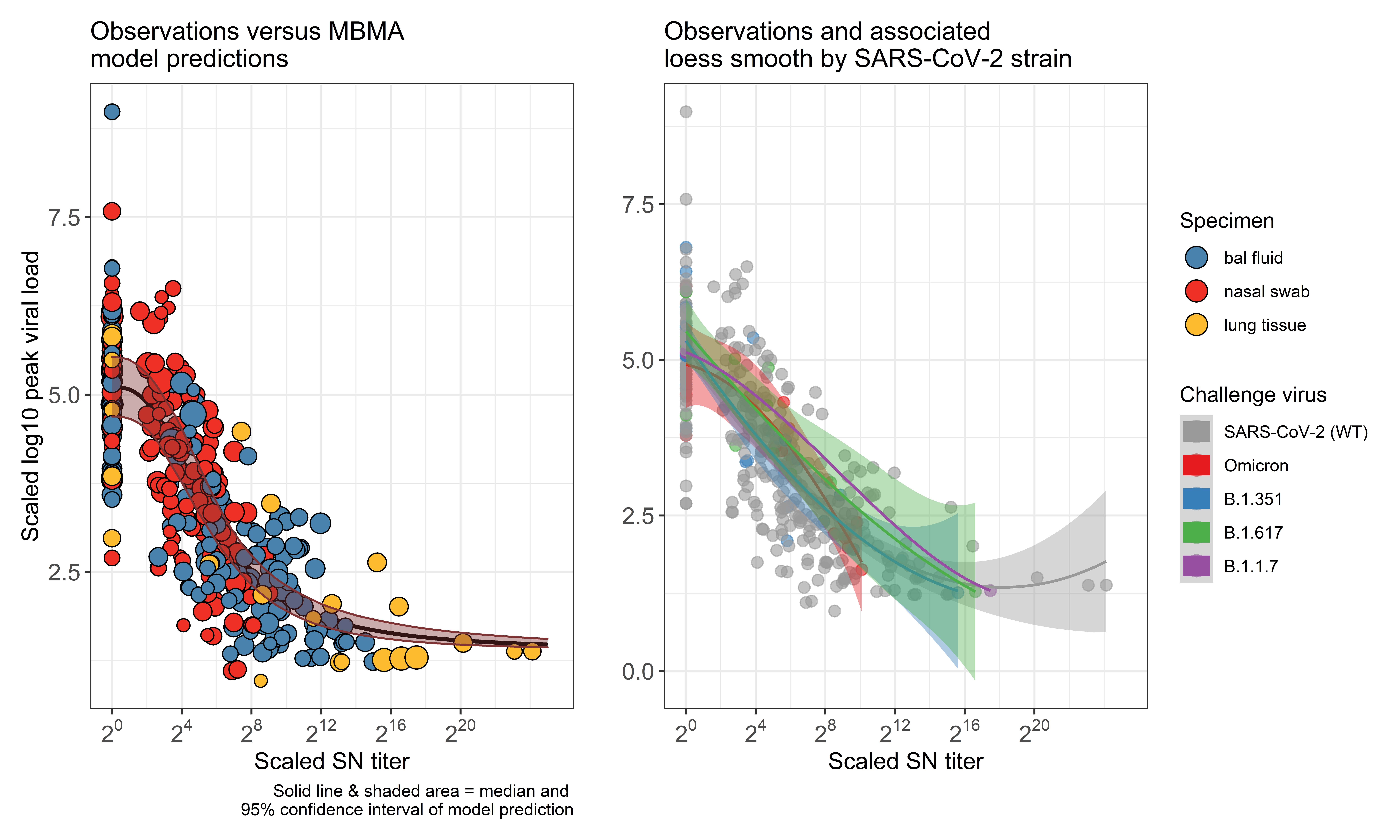
Methods: Previously, based on a systematic literature review, we performed a translational Model-Based Meta-Analysis (MBMA)1 integrating data of wild-type (WT) SARS-CoV-2 from 13 rhesus macaques (RM) studies and 8 clinical trials. The model is here updated with data from 32 additional RM studies including newer strains of SARS-CoV-2 (e.g., omicron). Non-linear mixed-effects modeling was used to quantify the relationship in RM between serum neutralizing (SN) titers after vaccination and peak viral load (VL) post challenge in relevant tissue matrices.
Results: The plot2 shows the model describes the relationship between SN titers and peak VL across all specimens well. The overlap between the confidence intervals across virus strains suggests that the model can be leveraged to describe RM data across viral SARS-CoV-2 strains.
Conclusion: The previous work1 demonstrated that RM VL is quantitatively predictive of clinical efficacy, and so this update provides a framework to predict clinical vaccine efficacy against newer variants using only RM data.
Kandala, B et al. Accelerating model-informed decisions for COVID-19 vaccine candidates using a model-based meta-analysis approach. eBioMedicine. 2022. [Accepted]
Lommerse J, Plock N, Cheung SYA, Sachs JR. V2ACHER: visualization of complex trial data in pharmacometric analyses with covariates. CPT Pharmcometrics.Sys.Pharmacol.10,1092-1106 (2021).
Relationship between SN titers and peak Viral Load in RM across all specimens and strains quantified via the MBMA model.